Posted on February 26, 2019 in ASRC News, Featured News, Structural Biology Initiative
The discovery could help in the development of novel, more effective antibiotics
NEW YORK, February 25, 2019 – From the complex to the simple, all life forms have mechanisms for translating environmental cues into cellular behavior that helps them survive. This universal activity may hold the key to understanding how common bacteria transform into virulent, deadly infections in humans, but the multifaceted protein sensing and signaling processes that allow lifeforms to adapt are poorly understood in even the simplest bacteria. A newly published paper in PNAS details how researchers at the Advanced Science Research Center (ASRC) at The Graduate Center of The City University of New York are using blue light and bacteria called Erythrobacter litoralis to unlock previously unknown details of one of these processes. Their findings could ultimately help drug developers create novel, more effective antibiotics and antifungal treatments.
E. litoralis are ocean-dwelling bacteria whose cellular behavior is stimulated when they come in contact with blue light. “We liked these bacteria as a study model because their response to blue light uses fewer steps than many other bacterial processes,” said first author Igor Dikiy, a National Institutes of Health (NIH) National Research Service Award postdoctoral fellow with the ASRC’s Structural Biology Initiative. “We believe some of the more complex, pathogenetic bacteria that live in the gut may also take their cues from light stimulation, so studying this light-sensing process in E. litoralis is a good place to begin understanding how such bacteria ‘turn on’ the ability to infect us.”
Methodology
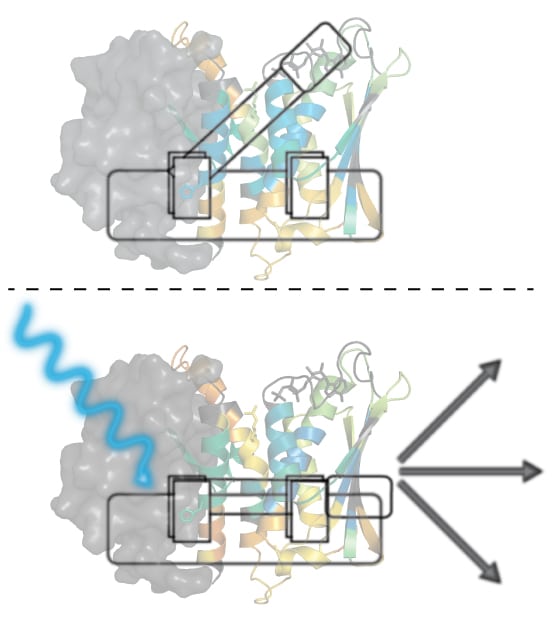
With support from the NIH and Burroughs Wellcome Fund, the researchers designed their study to elucidate how cellular activity is initiated and carried out within E. litoralis bacteria in response to light. Previous studies conducted in the lab of ASRC Structural Biology Initiative Director Kevin Gardner had already sorted out some of this process. Researchers knew, for example, that when blue light hits E. litoralis bacteria, it is detected by the sensory-domain portion of a histidine kinase protein called EL346. How that signal was next transmitted between this light-sensitive sensory domain and the catalytic kinase domain (or activity site) remained an open question. Did it go through a direct path like a telephone line, or something more unusual?
To answer this question, ASRC researchers employed two technologies that allowed them to track and determine what motions within the protein molecule are involved in communicating the sensing and signaling process. Using equipment at the ASRC Biomolecular Mass Spectrometry Facility and a process known as hydrogen-deuterium exchange by mass spectrometry [HDX-MS], researchers were able to detect changes in the EL346 protein’s mass after exposure to blue light. Additionally, working together with researchers at Cornell University’s National Biomedical Center on Advanced ESR Technologies (ACERT), the team used electron spin resonance (ESR) to measure changes in the distance between the protein’s sensory and catalytic domains after exposure to blue light. Together, these measurements allowed them to draw a picture of the protein’s internal control mechanisms.
Significance
“The appeal of these findings is that this particular sensing-signaling structure is unique to bacteria, so that means we can build off of these results to design new drugs that will only affect related signaling processes in the bacteria we’re targeting,” said Gardner.
The lab’s next goal is investigating how E. litoralis bacteria cells further respond to the blue light stimulus. “The ASRC is one of the few facilities in the area able to conduct this research because of our outstanding combination of expertise, instrumentation, and focus on interdisciplinary research approaches to tackle challenging problems like this,” added Gardner.
###
Organizational Attribution
Our correct name is the Advanced Science Research Center at The Graduate Center of The City University of New York. For the purpose of space, Advanced Science Research Center, GC/CUNY is acceptable. On second reference, ASRC is correct.
About the Advanced Science Research Center
The ASRC at The Graduate Center elevates scientific research and education at CUNY and beyond through initiatives in five distinctive, but increasingly interconnected disciplines: environmental sciences, nanoscience, neuroscience, photonics, and structural biology. The ASRC promotes a collaborative, interdisciplinary research culture with renowned researchers from each of the initiatives working side by side in the ASRC’s core facilities, sharing equipment that is among the most advanced available.
About The Graduate Center of The City University of New York
The Graduate Center of The City University of New York (CUNY) is a leader in public graduate education devoted to enhancing the public good through pioneering research, serious learning, and reasoned debate. The Graduate Center offers ambitious students more than 40 doctoral and master’s programs of the highest caliber, taught by top faculty from throughout CUNY — the nation’s largest public urban university. Through its nearly 40 centers, institutes, and initiatives, including its Advanced Science Research Center (ASRC), The Graduate Center influences public policy and discourse and shapes innovation. The Graduate Center’s extensive public programs make it a home for culture and conversation.
###
