Posted on September 18, 2024 in ASRC News, Structural Biology Initiative
The approach can aid in developing new drug therapies.
Detecting how and where small molecules bind to proteins is challenging and time-consuming, but essential for determining how a protein can be targeted by drugs. PTP1B, a protein linked to diabetes and cancer, is difficult to target because most molecules that bind to it also bind to protein relatives of PTP1B, resulting in undesired side effects. Discovering more ligands and binding sites on a particular protein can create new opportunities for drug research aimed at treating specific diseases.
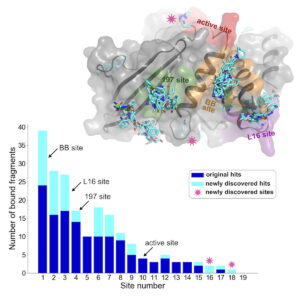
Using a new computational analysis technique, researchers at the CUNY ASRC analyzed X-ray crystallography data and discovered 65 new small-molecule fragments that bind to PTP1B. The results of their analysis have been published in a recent issue of the journal Structure.
“We use something called crystallographic fragment screening, in which we expose proteins to a lot of so-called small-molecule fragments and shoot X-rays at them,” said the study’s principal investigator Daniel Keedy, a researcher with the CUNY ASRC’s Structural Biology Initiative and a professor of chemistry and biochemistry at City College of New York. “From the resulting X-ray diffraction data, we determine the fragments that bind. The idea is that you can get a bunch of these fragments and then see how the puzzle pieces fit together to form a drug that works on that protein.”
Previous studies used a technique called PanDDA to identify some molecules that bind to PTP1B. PanDDA looks for deviations across datasets that indicate a ligand is binding to the structure. The newly developed computational technique, called cluster4x, improves the success rate of finding bound ligands, resulting in the discovery of 65 new instances of ligands binding to the protein — a 50% increase over previous results. While most new ligands bound to already known sites, new binding sites were also discovered.
One surprising finding was that some fragments caused structural changes in the protein far from where the small molecules bind. This suggests that targeting these sites could influence the enzyme’s function, offering new avenues for drug design.
“A lot of ligands just bind and don’t really do anything to the protein,” said Keedy. “But in one of these cases, we saw a ligand bind to one site that caused a series of change in the protein structure leading to the active site, kind of like a domino effect. We call this allostery, or action at a distance. Ligands that have these allosteric properties are very interesting because you could develop an allosteric drug from them that would avoid the problems with active-site drugs.”
The newly discovered PTP1B structures provide valuable data for understanding protein-ligand interactions, which is useful for developing new drugs. In future studies, the team will explore how to make the new ligands bind more tightly to the new protein sites and use cluster4x for fragment screening of other proteins involved in diseases such as Alzheimer’s and Parkinson’s.
The research team includes Helen Ginn, head of the Protein Machinists Lab at the University of Hamburg, who developed the computational method cluster4x, and Tamar Mehlman, a postdoctoral researcher in the Keedy lab who applied the new technique to the datasets.
